By A Mystery Man Writer
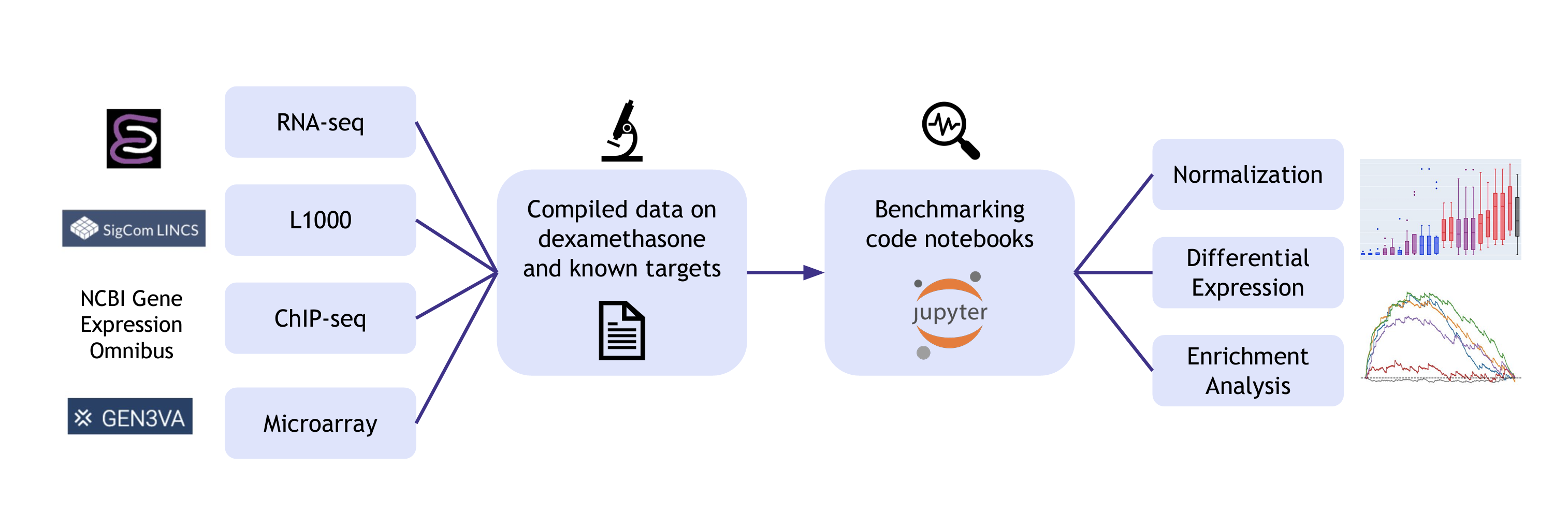
The Dex-Benchmark resource – datasets and code to evaluate algorithms for transcriptomics data analysis
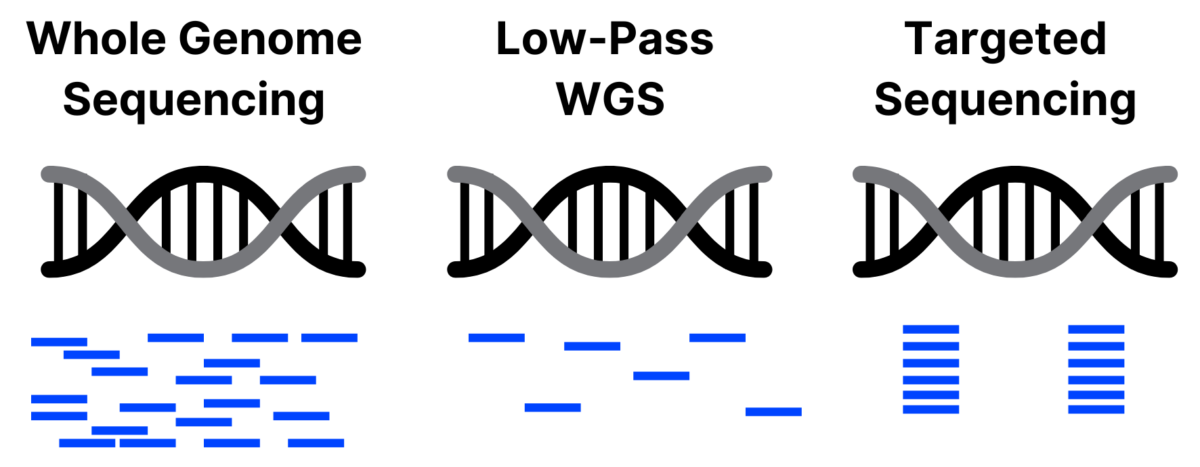
Low-Pass Whole Genome Sequencing
Frontiers Evaluation and Recommendations for Routine Genotyping Using Skim Whole Genome Re-sequencing in Canola

scIALM: A method for sparse scRNA-seq expression matrix imputation using the Inexact Augmented Lagrange Multiplier with low error - Computational and Structural Biotechnology Journal
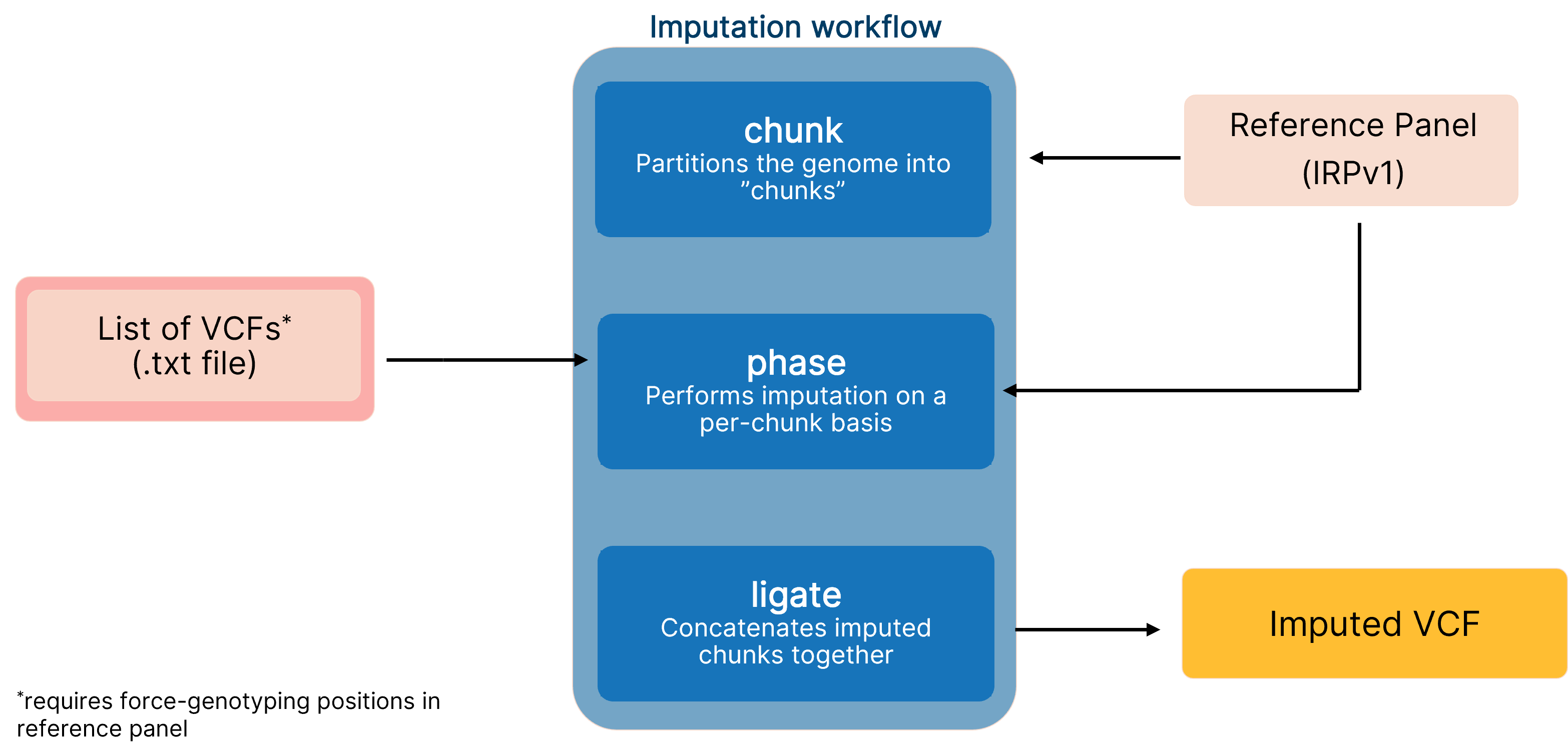
Boosting variant calling performance using a high-quality reference panel for imputing low-coverage sequencing data

PDF) Evaluating the concordance between sequencing, imputation and microarray genotype calls in the GAW18 data
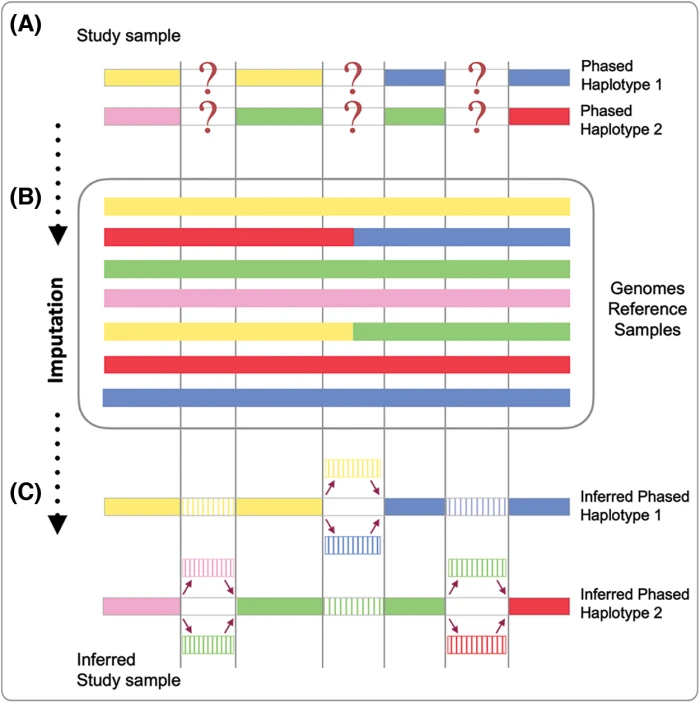
BIOCELL, Free Full-Text

Best practices for analyzing imputed genotypes from low-pass sequencing in dogs

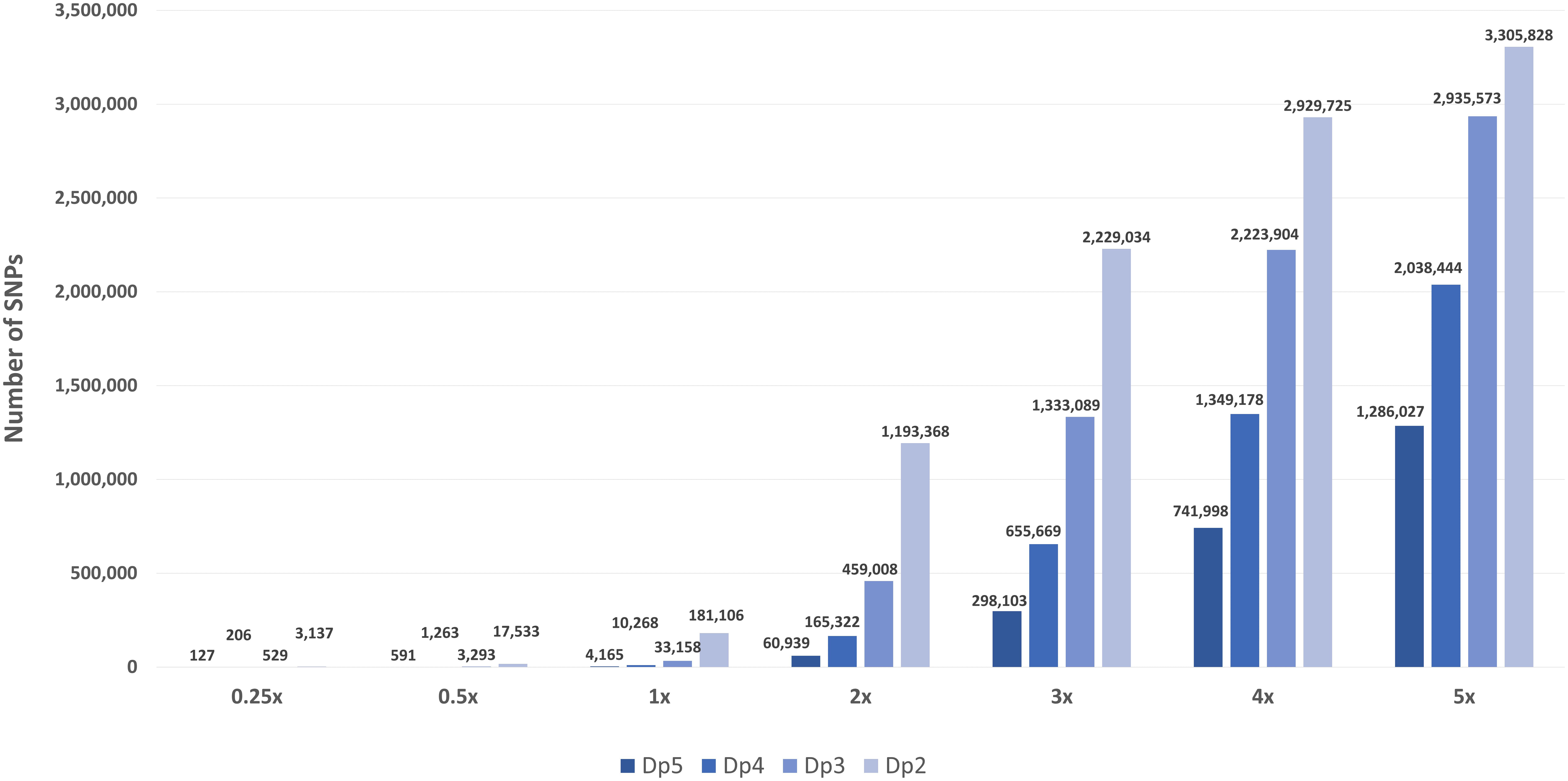
Frontiers Ultra Low-Coverage Whole-Genome Sequencing as an Alternative to Genotyping Arrays in Genome-Wide Association Studies
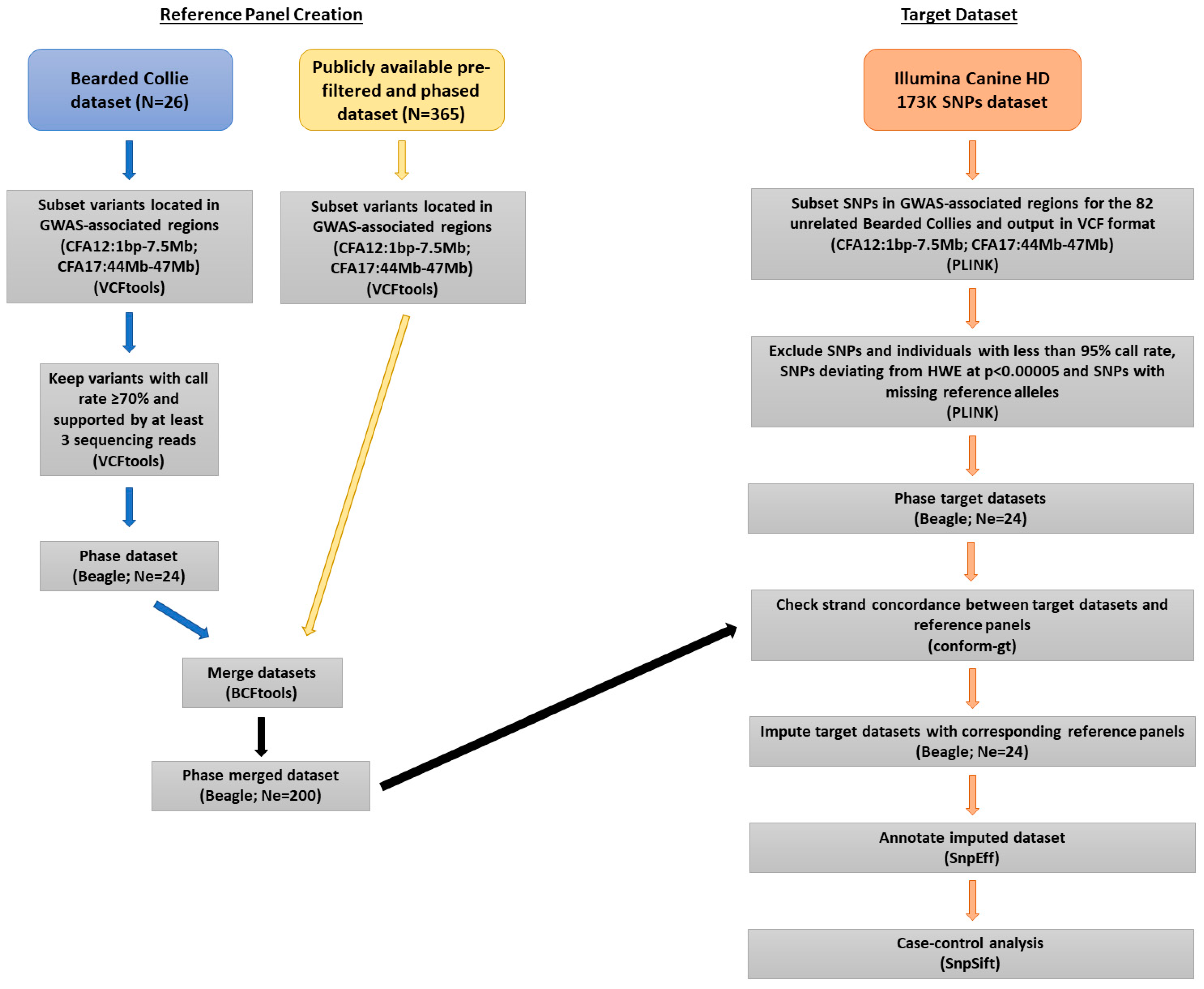
Genes, Free Full-Text

Evaluating genotype imputation pipeline for ultra-low coverage ancient genomes